Abstract
Acute myeloid leukemia (AML) is a clinically heterogeneous disease characterized by chromosomal abnormalities, gene mutations, and inter- and intra-patient genetic and phenotypic variability. Therapies specifically targeting cell surface antigens are promising treatments for AML, but surface antigen expression variability and potential antigen escape mechanisms may impact efficacy of single-targeting therapies. Delineating surface antigen expression uniformity and density between patients and during disease progression from diagnosis to relapse will enable rational prioritization of AML antigens for adaptive and combinatorial targeting. Here, we elucidate surface antigen expression patterns for candidate therapeutic targets by performing multimodal integrative analysis of AML surface proteins and transcriptomes of matched diagnosis and relapse clinically annotated samples.
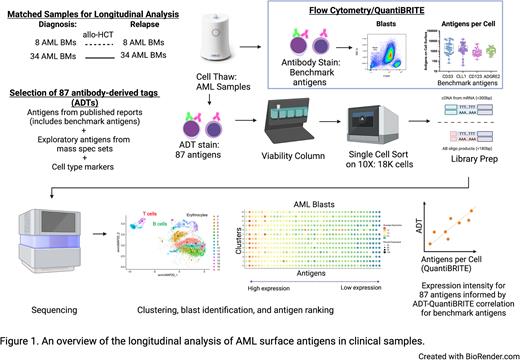
To identify potential therapeutic AML targets, we compiled a comprehensive list of protein surface targets reported in clinicaltrials.gov, literature, and large proteomics mass spectrometry datasets of AML patient bone marrow samples (deBoer et. al. Cancer Cell 2018; Jayavelu et al, Cancer Cell 2022). To focus on surface antigens and reduce the risk of targeting antigens that may have off-tumor solid tissue toxicity, we then excluded antigens that lack plasma cell membrane localization or are expressed on healthy non-hematologic tissues. We adapted Cellular Indexing of Transcriptomes and Epitopes by Sequencing (CITE-seq) to generate longitudinal single-cell multimodal readouts of both the transcriptome and targeted surface proteins of matched diagnostic and relapsed samples from 42 adults with AML (Figure 1). Antibody derived tags (ADTs) against selected antigens were used at saturating conditions. Single cell libraries were prepared with 10X Genomics 5' Feature Barcoding Library Kit v2 and sequenced on Illumina NextSeq 2000. Cellranger v6.0.0 and Seurat v3 were used for mapping, read quantification, and analysis, respectively. We leveraged AML cell lines with known expression of benchmark antigens to assess the accuracy of the ADT readout by correlating it with antibody bound per cell (ABC) values derived from the traditional flow cytometry-based QuantiBRITE assay.
Patient bone marrow data were projected onto low dimensional space using UMAP to visualize the population structure of the data. We observed separation of AML blasts from healthy cell lineages and delineation of blast gating based on CD45 expression. k-nearest neighbors clustering identified phenotypically diverse clusters within AML blasts within each patient sample, enabling quantification of specific antigen-positive clusters and antigen co-expression patterns. We found strong correlation between ADT readout and corresponding ABC values for cell surface protein expression on AML blasts of our benchmark antigens (CD33, CD123, CLL1, and ADGRE2). We then proceeded to systematically rank 87 AML surface antigens in blast populations based on ADT readout. Moreover, transcriptome profiling allowed investigation of antigens with limited antibody availabilities, exploration into novel cell states, and identification of signaling pathways that may associate with the presence of particular surface antigens. Identification and analysis of leukemic stem cells are ongoing. Healthy bone marrow and peripheral blood will be analyzed alongside AML samples to elucidate antigen target expression during normal hematopoiesis.
In summary, our multimodal analysis of AML surface antigens informs cell surface clonal heterogeneity of patient AML and builds a reference atlas of expression patterns and intensities of potential surface AML antigen targets. In ongoing work, we are analyzing correlations between clonal surface antigen expression in longitudinal analysis of diagnosis and relapse samples as well as correlation with clinical parameters. This comprehensive surface antigen expression profiling will inform selection of therapeutically targetable and consistently expressed antigens to address AML heterogeneity in future immunotherapy targeting studies.
Note: J.E. and M.U. share co-first authorship.
Disclosures
Etchin:Vor Biopharma: Current Employment, Current equity holder in publicly-traded company. Ung:Vor Biopharma: Current Employment, Current equity holder in publicly-traded company. Halfond:Vor Biopharma: Current Employment, Current equity holder in publicly-traded company. Keschner:Vor Biopharma: Current Employment, Current equity holder in publicly-traded company. DiFazio:Vor Biopharma: Current Employment, Current equity holder in publicly-traded company. Mundelboim:Vor Biopharma: Current Employment, Current equity holder in publicly-traded company. Ge:Vor Biopharma: Current Employment, Current equity holder in publicly-traded company. Lydeard:Vor Biopharma: Current Employment, Current equity holder in publicly-traded company. Chakraborty:Vor Biopharma: Current Employment, Current equity holder in publicly-traded company.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal